MiSynPat: an integrated knowledge base linking clinical, genetic, and structural data for disease-causing mutations in human mitochondrial aminoacyl-tRNA synthetases.
Moulinier L, Ripp R, Castillo G, Poch O, Sissler M.
Human Mutation 2017 Jun 12. doi: 10.1002/humu.23277. PMID: 28608363
MiSynPat is a web-based application and knowledgebase dedicated to human mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs) and clinically relevant (disease-related) mutations.
It is dedicated to a broad audience of specialists (clinicians and researchers) and of non-specialists (for instance patients and patient associations,
but also anyone willing and interested)
Misynpat provides a dynamic, comprehensive and ergonomic graphical user interface with two main applications:
- A Database of existing mutations providing the exhaustive list of mutations, links to literature, information, indicating allelic compositions in patients, depicting the mutations on protein sequences, multiple sequence alignments and 3D modeled structures.
- A Web application that offers tools to visualize the impact of possible new mutation on sequence-conservation-structure interrelation of mt-aaRSs for potential help to diagnosis.
Aminoacyl-tRNA synthetases (aaRSs) are enzymes implicated in one of the key steps of the protein biosynthesis process. They are responsible for the specific ligation of amino acids to their cognate tRNAs via a two-steps reaction (amino acid activation and transfer of the activated amino acid onto the cognate tRNA). In human mitochondria, mt-aaRSs are necessary for the synthesis of 13 mt-DNA-encoded proteins, all subunits of the respiratory chain complexes, and thus, implicated in the synthesis of ATP, the major source of cellular energy. Human mt-aaRSs are all encoded by nuclear genes, by a set of genes distinct from those coding for the cytosolic aaRSs, with solely two exceptions, the GlyRS and the LysRS. Furthermore, up to now, no gene coding for the mt-GlnRS has yet been found and Gln-tRNAGln is produced via an alternate indirect pathway.
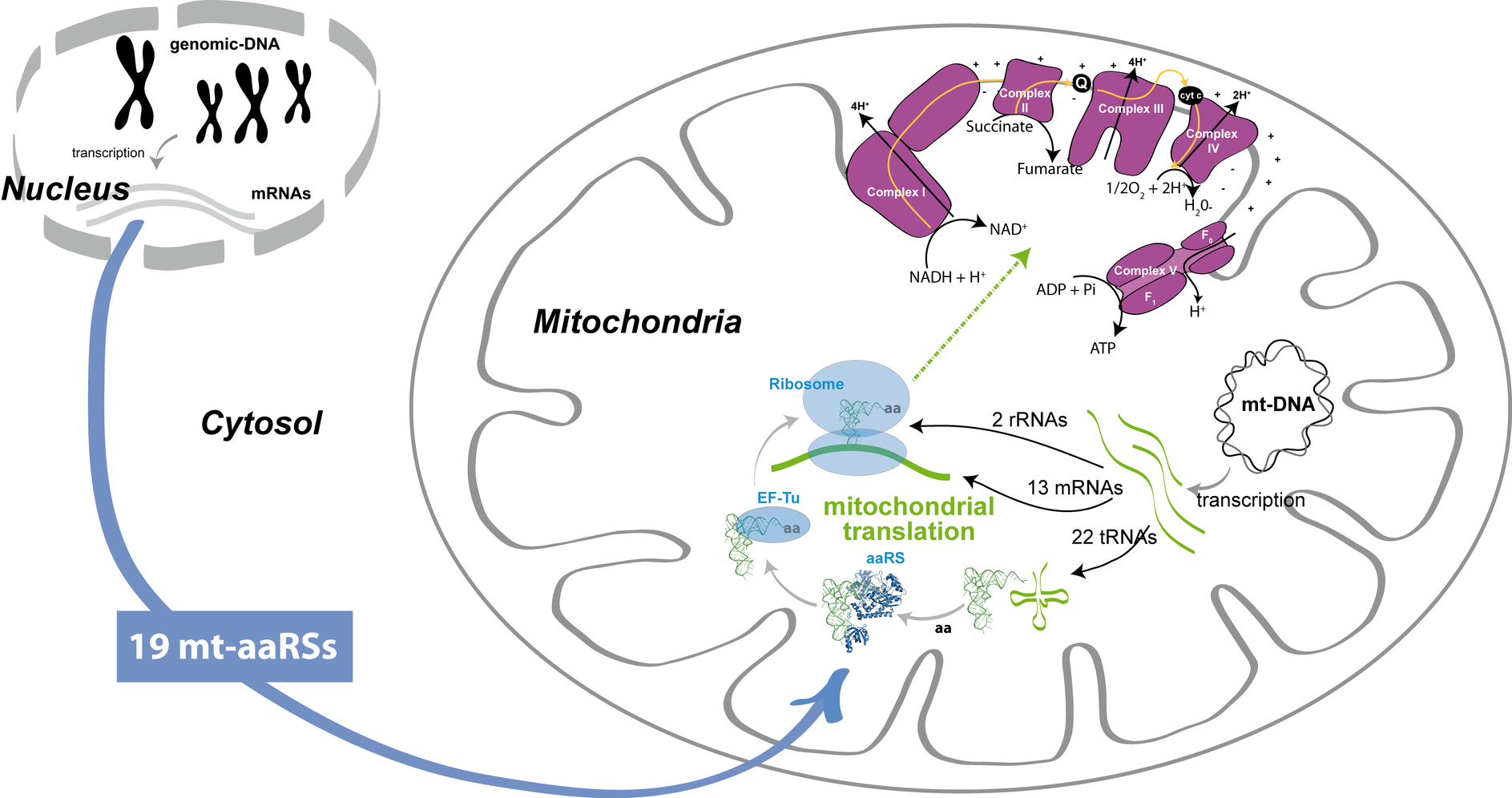
Human mt-aaRSs and mitochondrial translation: Under normal conditions, mt-aaRSs participate in the synthesis of the 13 mt-DNA encoded subunits of the respiratory chain complexes, and are thus implicated in the cellular energy production in the form of ATP. RNA components (i.e. 22 tRNAs, 13 mRNAS and 2 rRNAs) for the mitochondrial translation machinery are encoded by the mitochondrial genome (mt-DNA). Conversely, all requested proteins are encoded within the nuclear genome, translated into the cytosol, and imported into the mitochondria. This is for instance the case for the 19 aaRSs of mitochondrial location. As mentioned, ther is no gene coding for a human mt-GlnRS.
The first correlation between a mutation affecting a mt-aaRS and a human disease dates back to the year 2007, when mutations within the DARS2 gene, coding for mt-AspRS, were associated with Leukoencephalopathy with Brain stem and Spinal cord involvement and Lactate elevation (LBSL). Many other mutations have been discovered since then, so that nowadays 17 out of the 19 genes encoding mt-aaRSs have been reported to be impacted by pathogenic mutations. All mutations reported so-far lead to autosomal (i.e. affecting any chromosome other than a sex chromosome) recessive disorders. Patients are either compound heterozygotes (with both recessive alleles for a same gene mutated at different location) or homozygotes (with both recessive alleles for a same gene identically mutated at the same location). As new mutations are recurrently described, MiSynPat offers follow-up tools of pertinent information, allying literature and sequence-structure-conservation knowledge, so that to assist data interpretation in pursuance of reaching a lucid diagnosis.